Bucketing
An NMR spectrum may contain several thousands of points, and therefore of variables. In order to reduce the data dimensionality binning is commonly used. In binning the spectra are divided into bins (so called buckets) and the total area within each bin is calculated to represent the original spectrum. The more simple approach consists to divide all the spectra with uniform areas width (typically 0.04 ppm). Due to the arbitrary division of peaks, one bin may contain pieces from two or more peaks which may affect the data analysis. We have chosen to implement the Adaptive, Intelligent Binning method [De Meyer et al. 2008] that attempt to split the spectra so that each area common to all spectra contains the same resonance, i.e. belonging to the same metabolite. In such methods, the width of each area is then determined by the maximum difference of chemical shift among all spectra.
How to proceed (see fig below)
- Specify a relevant zone in order to estimate the noise level.
- Choose a resolution factor between 0.1 and 0.6 (0.5 is the default value); the smaller value the greater resolution
- Select one or more PPM zones for applying the binning and put them in the box of ‘PPM ranges’
- Choose the threshold of the Signal/Noise Ratio (SNR) so that the buckets having a lower average integration will be excluded.
- Then launch
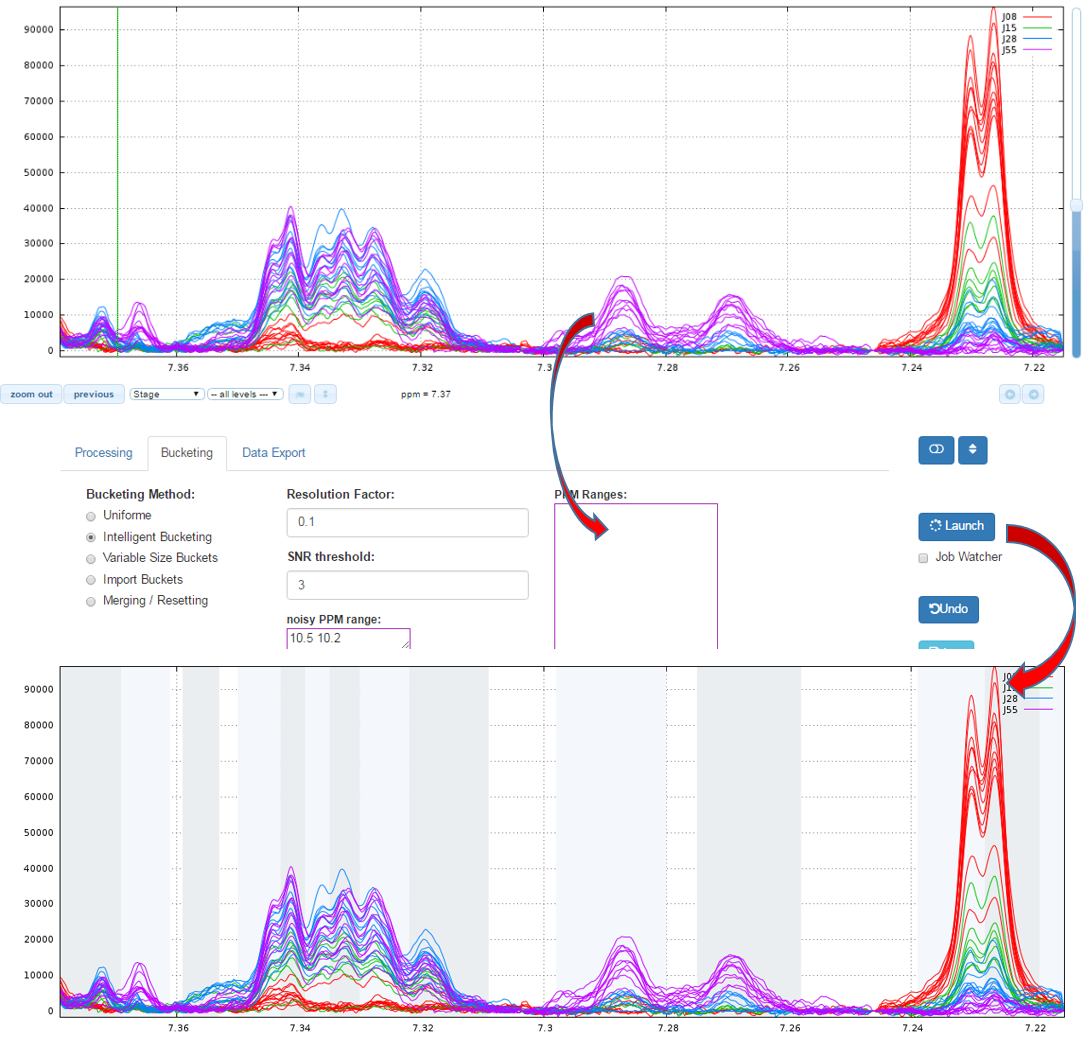
Warnings:Do not choose a resolution parameter too small in an area where alignment has not been made. NMRProcFlow makes it possible to adapt area by area the right resolution.
de Meyer T, Sinnaeve D, van Gasse B, Tsiporkova E, Rietzschel E, de Buyzere M, Gillebert T, Bekaert S, Martins J, van Criekinge W (2008) NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Anal Chem 80:3783–3790

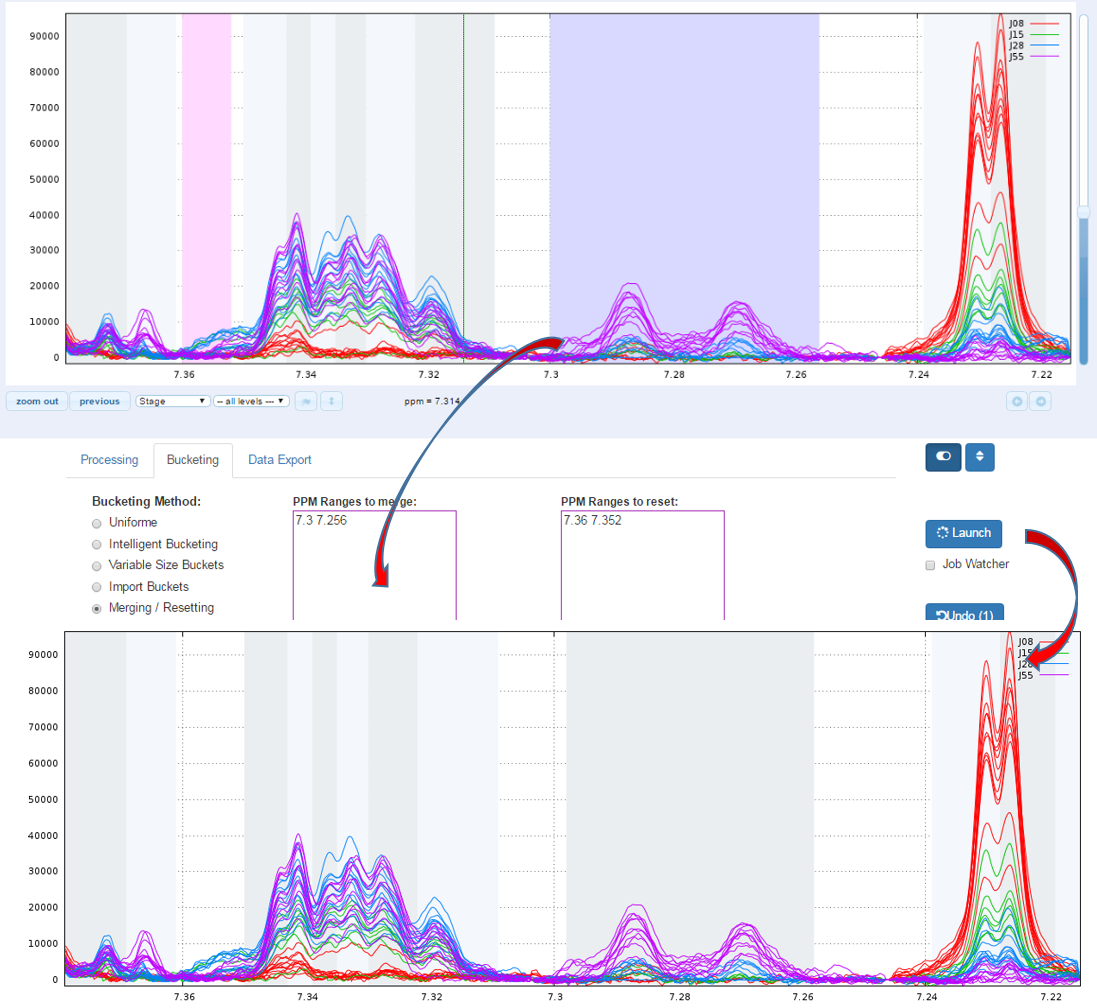
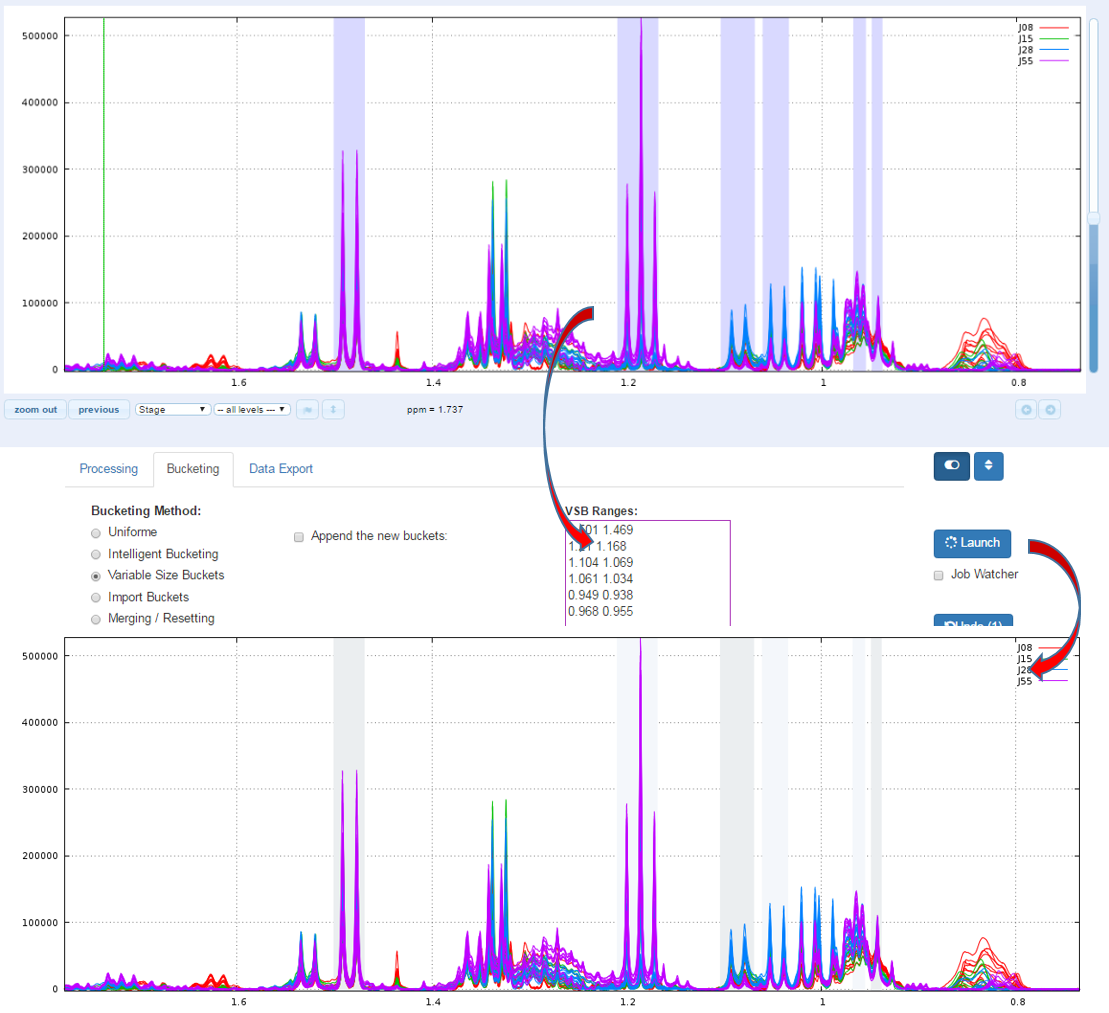
Another way for obtain buckets is to choose yourself ppm ranges you want to integrate. This method is typically used for the Targeted metabolomics approach where only few peaks corresponding to targeted compounds are selected whose their size is depending of the signal pattern.

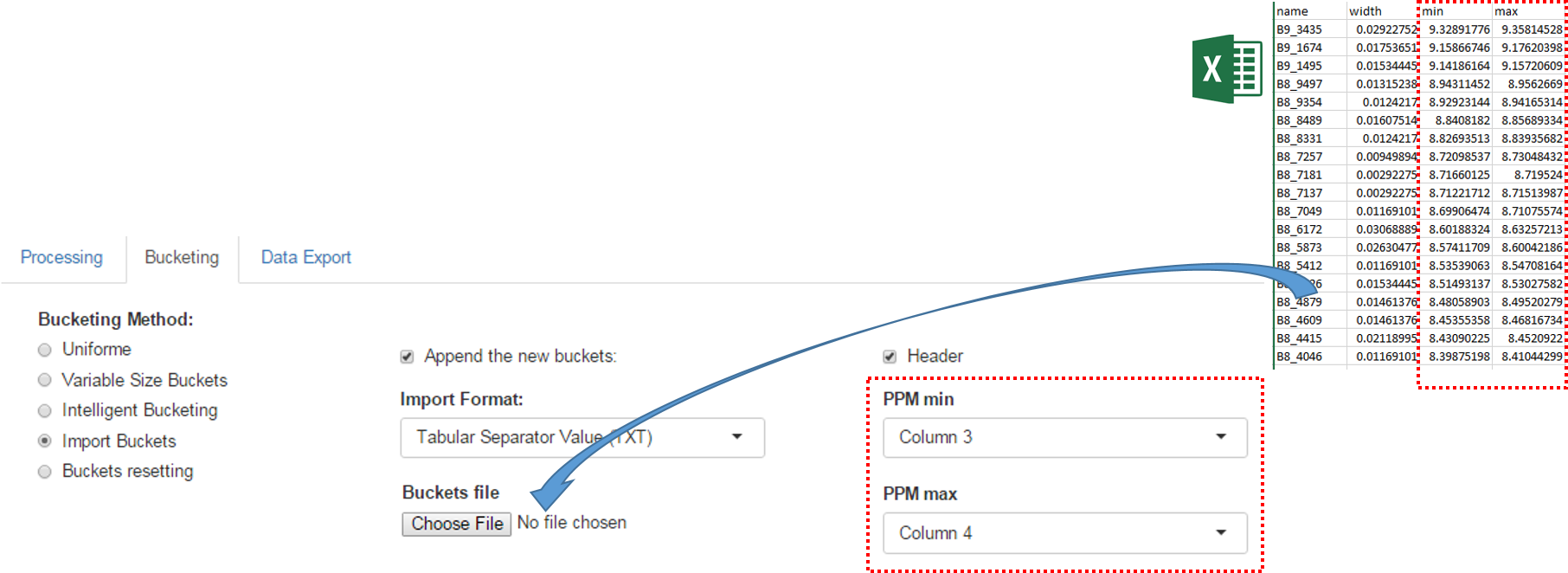
An exported buckets table can be imported into NMRProcFlow in order to retrieve the same bucketing obtained in a previous work session
How to proceed (see fig below)
- Choose the format corresponding of the imported file
- Check if the imported file have an header line or no
- Choose the corresponding columns for the lower and upper ppm bounds of the buckets.
- Check if imported buckets will be append to those already defined or not. If not, all buckets previously defined will be erased.
- Then launch