Batch mode execution
NMRProcFlow allows experts to build their own spectra processing workflows, in order to become models applicable to similar NMR spectra sets, i.e. stated as use-cases.
In the Replay the same processing workflow section, we have seen how to export a macro-commands file in order to be replayed in the NMRProcFlow GUI, i.e. in interactive mode. Now the idea as depicted in the figure below, is to consider the macro-commands file as a processing model that can be applied on other subsets and replayed in batch mode.
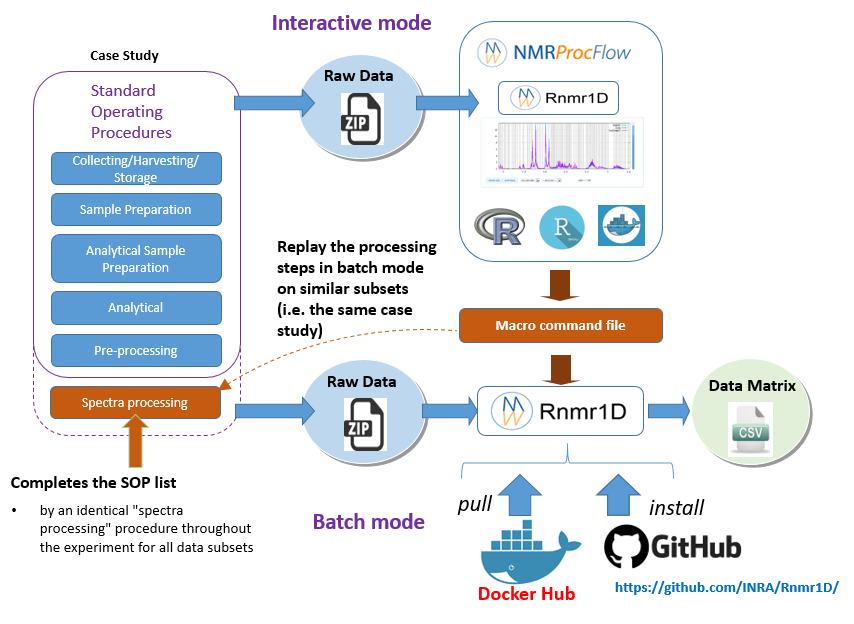
By extension, we consider the implementation of NMR spectra processing workflows executed in batch mode as relevant provided that we want to process in this way very well-mastered and very reproducible use cases, i.e. by applying the same Standard Operating Procedures (SOP).
- Interactive mode: A subset of NMR spectra is firstly processed in interactive mode using NMRProcFlow having a NMR spectra viewer to allow the expert eye to disentangle the intertwined peaks, in order to build a well-suited workflow. This mode could be also named the 'expert mode'.
- Batch mode: Then, other subsets that could be regarded as either similar or being included in the same case study, can be processed in batch mode (See below the two options)
* To run the R Rnmr1D package within a R session, go to the github repository https://github.com/INRA/Rnmr1D. You can also see the online vignette that illustates the Rnmr1D package features
Cloud-based Metabolomics Data Processing
* To run the Rnmr1D application on a Cloud Research Environment (CRE), go to the Galaxy Portal of the EU PhenoMenal H2020 project.
* See the presentation "NMRProcFlow combined with the computing power of a Cloud Research Environment".