Data preparation phase
The current version of NMRProcFlow accepts raw data come from four major vendors namely Bruker GmbH, Agilent Technologies (Varian), Jeol Ltd and RS2D. Moreover, we also support the nmrML format (See nmrml.org for further information on this format)
Regarding Bruker, two types of raw data are accepted: Free Induction Decay (fid) and pre-processed raw spectra (1r). In both cases, the folder structure must follow that of the Bruker TopSpin software. "Pre-processed" means that it assumes that Fourier transform and phase correction have been applied on all spectra so that their corresponding processing directory (under 'pdata') exists along with their real spectrum (i.e 1r file). In the case where the input raw data are FID, the spectral pre-processing is automatically performed. See the Spectral pre-processing for 1D NMR section.
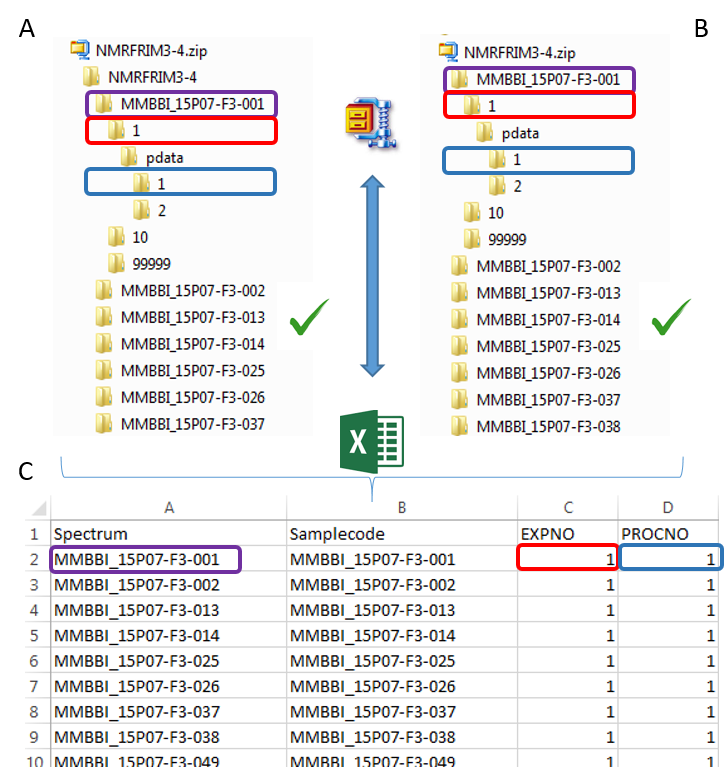
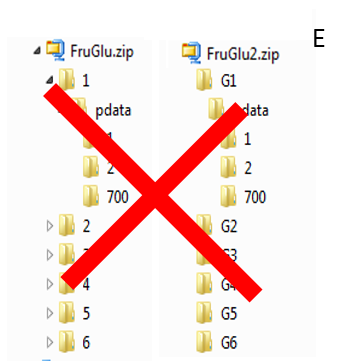
To ease the preparation phase, simply zip the entire directory including all spectra of the experiment. This means that it is useless to perform any prior selection or having to rename the numbers of experiment and processing. The figure below shows an example of ZIP file along with its corresponding samples file.
The colored boxes show the correspondences.
Supported directory structures for Bruker NMR spectra
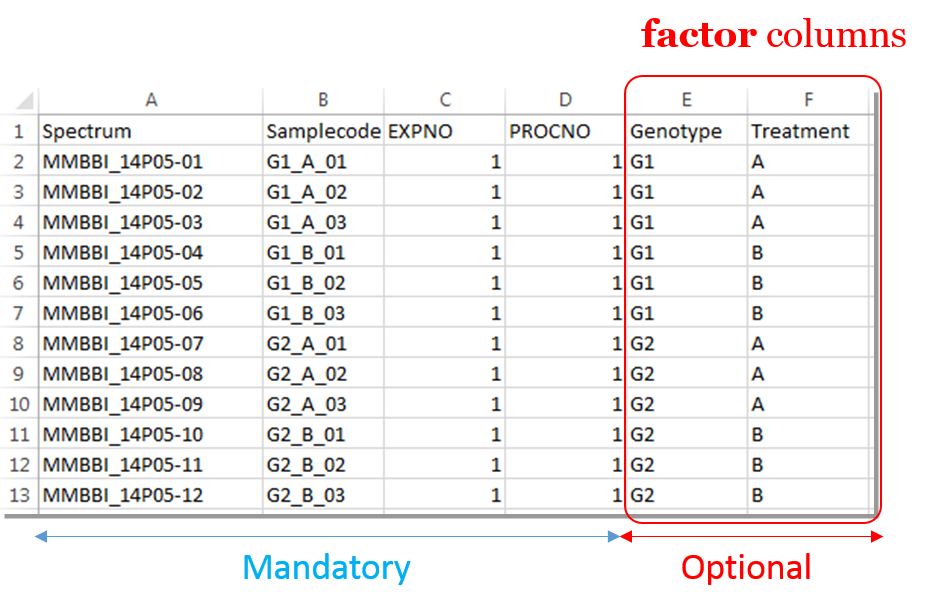
Information provided within the samples file must correspond to the directories contained in the ZIP file. (The colored boxes show the correspondences)
- The 'rawdata' column may include all directories or just a subset contained in the ZIP file.
- The 'Samplecode' column can be filled with the biological sample name or can be just a copy-paste of the 'Rawdata' column.
- The 'expno' and 'procno' columns correspond to the experiment number (i.e. FID) and the processing number (i.e. 1r) respectively.
- Several factor columns can be added which will allow spectra to be visualized according to theirs factor levels (see example online). The only constraints on factor names are that they must be alphanumeric characters [0..9, a..z, A..Z]. The underscore '_' is also allowed.
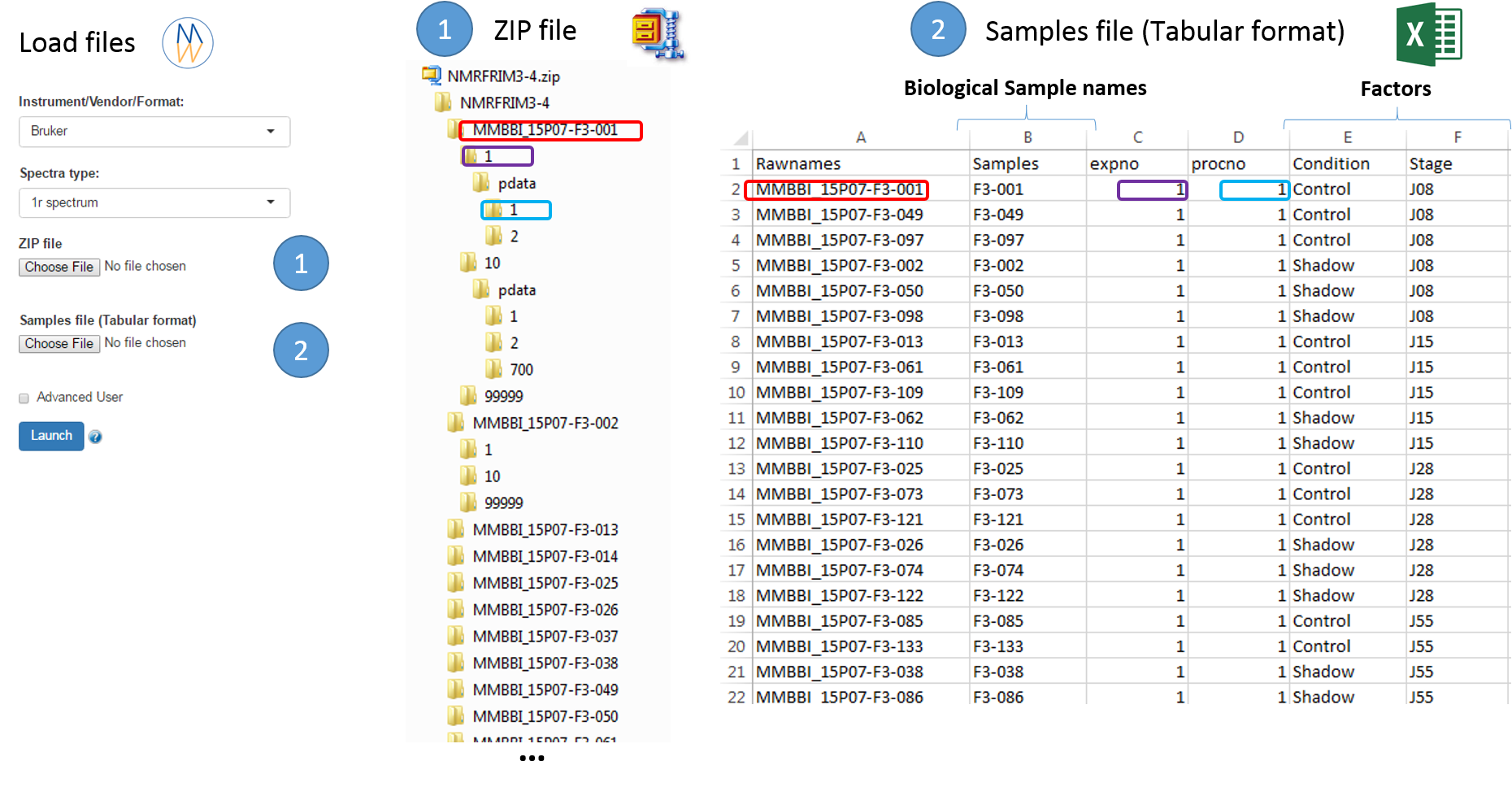
In this way, it becomes easy to select each NMR spectrum that we want to include into the spectra serial in order to be processed together. In the absence of the file of samples provided as an input, NMRProcFlow will consider all of the root directories in the zip file by default, looking for the smallest FID identifier (expno) and the smallest processing identifier (procno) for each of them.
Warnings: In the case where the input raw data are FID, the 'procno' column has to be kept (e.g. filled with zero) so that the samples file is thus compliant with both type of input data (fid & 1r)
To facilitate the generation of the samples file, it is possible to proceed as follows:
- Upload the ZIP file only, and then NMRProcFlow will produce a text file containing the acquisition and processing parameters for each spectrum taken into account by default,
- Download the parameters file ('Export Parameters'),
- Edit this file to serve as a starting template for generating the samples file.
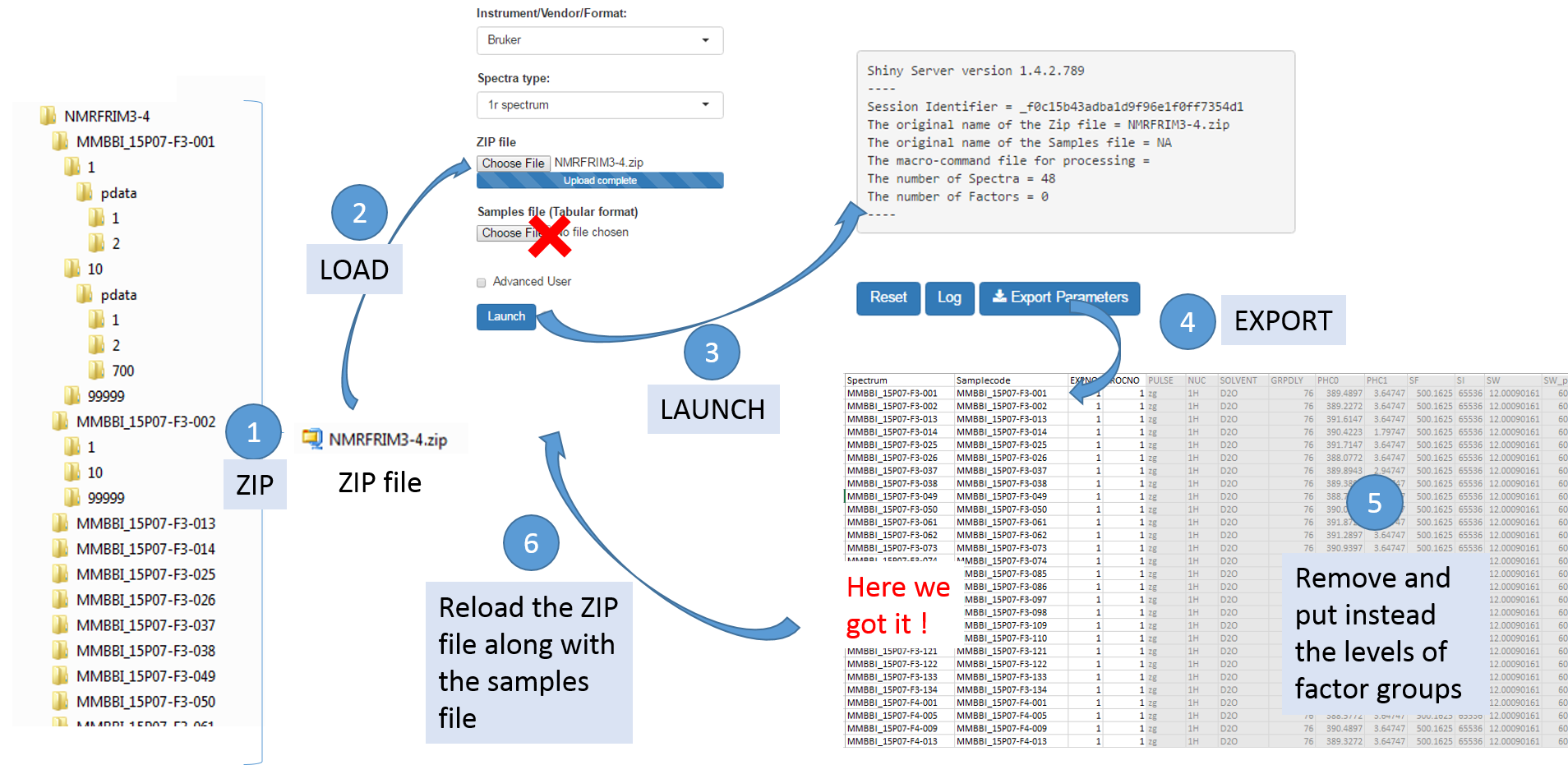
Once uploaded files, you can click on 'Launch' to start the pretreatment.
Once completed, the list of spectra considered with their acquisition and pre-processing parameters is provided
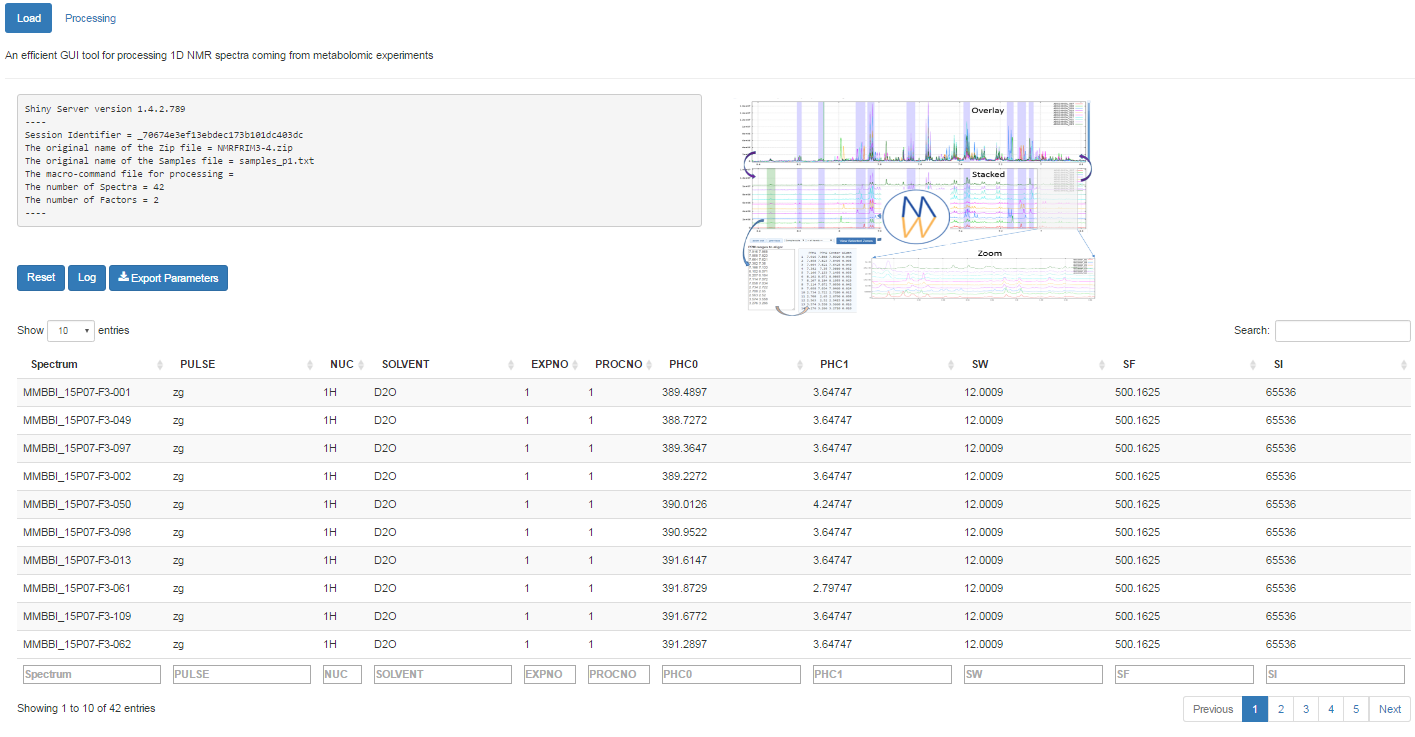
To switch to the processing steps, you must click on the 'Processing' tab at the top of the screen.

Regarding Agigent/Varian, only the Free Induction Delay are accepted, given that there is no normalized folder structure for pre-processed raw data provided by the OpenVnmrJ software. See the Spectral pre-processing for 1D NMR section to know how to choose parameters.
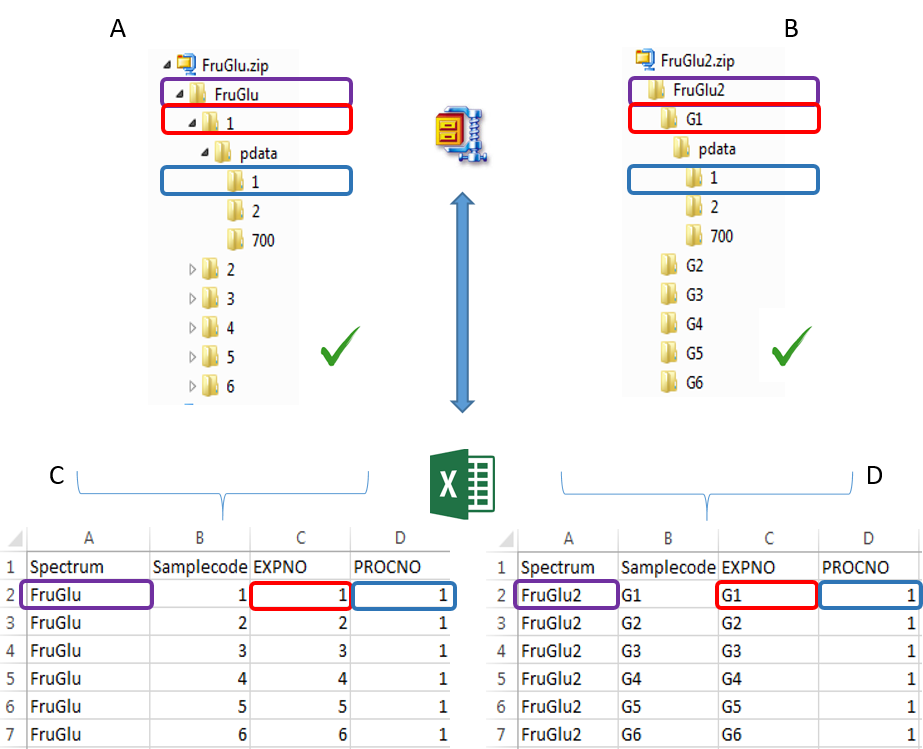
To ease the preparation phase, simply zip the entire directory including all spectra of the experiment. This means that it is useless to perform any prior selection or having to rename the numbers of experiment and processing. The figure below shows an example of ZIP file along with its corresponding samples file.
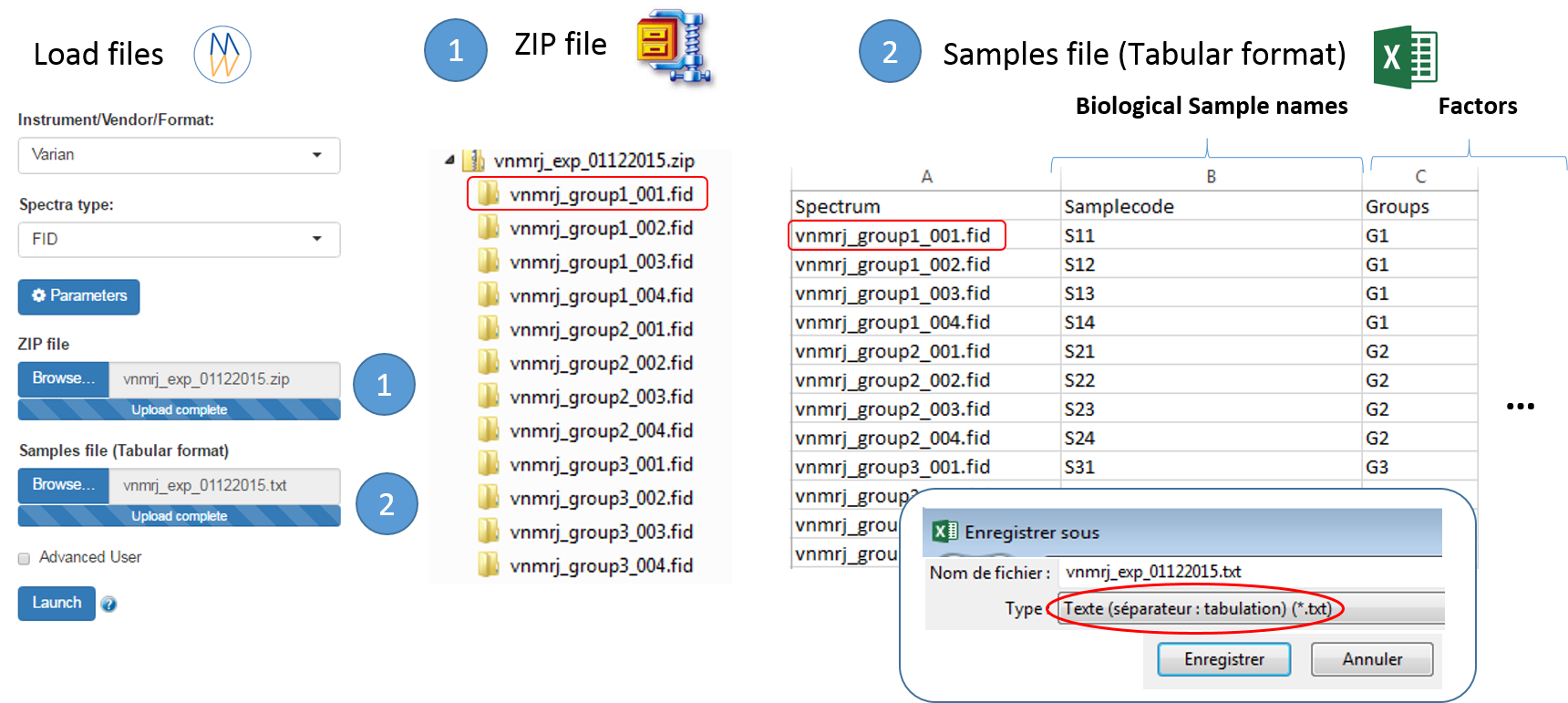
Information provided within the samples file must correspond to the directories contained in the ZIP file. (The colored boxes show the correspondences)
- The 'rawdata' column may include all directories or just a subset contained in the ZIP file.
- The 'Samplecode' column can be filled with the biological sample name or can be just a copy-paste of the 'Rawdata' column.
- Several factor columns can be added which will allow spectra to be visualized according to theirs factor levels. The only constraints on factor names are that they must be alphanumeric characters [0..9, a..z, A..Z]. The underscore '_' is also allowed.
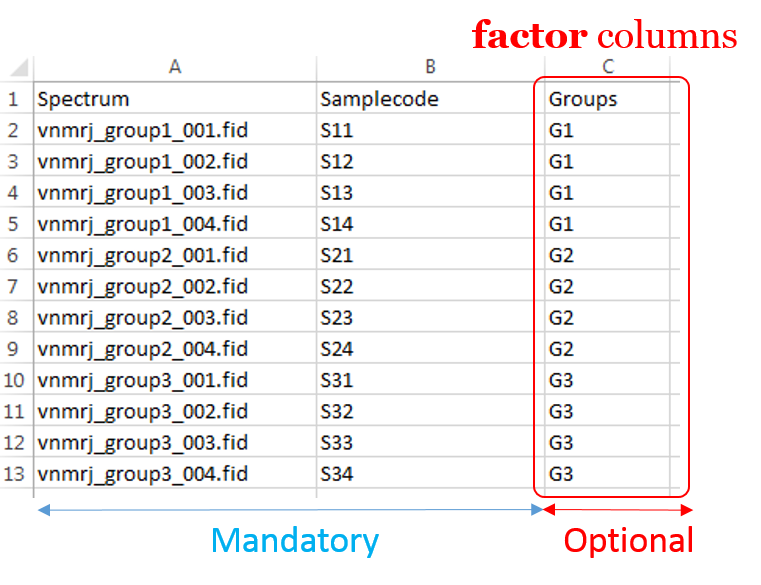
In this way, it becomes easy to select each NMR spectrum that we want to include into the spectra serial in order to be processed together. In the absence of the file of samples provided as an input, NMRProcFlow will consider all of the root directories in the zip file by default, looking for all FID files.
To facilitate the generation of the samples file, see the corresponding section in the 'Bruker' tab
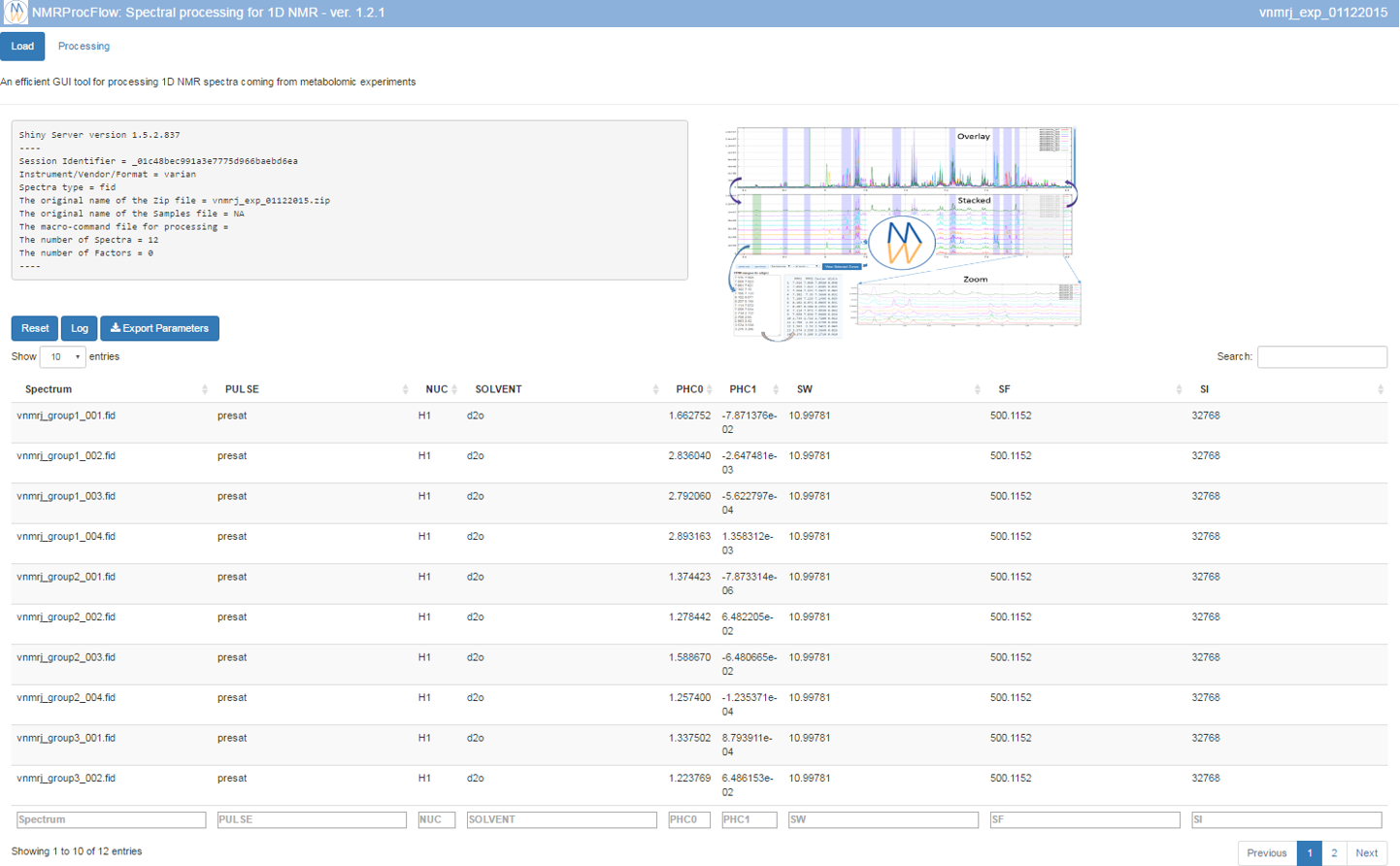
Once uploaded files, you can click on 'Launch' to start the pretreatment.
Once completed, the list of spectra considered with their acquisition and pre-processing parameters is provided
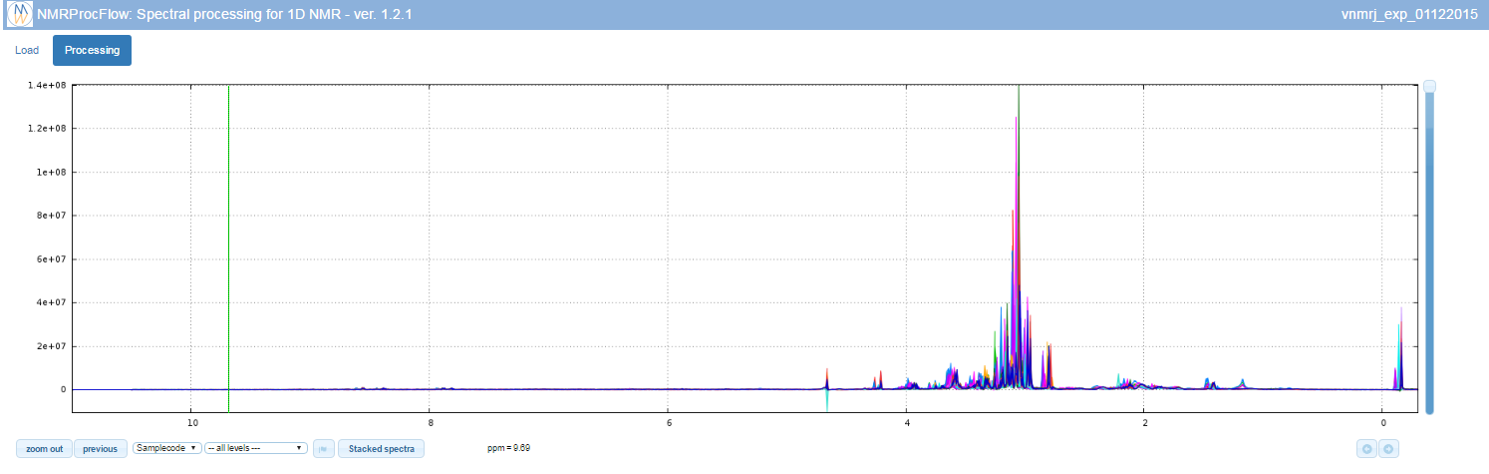
To switch to the processing steps, you must click on the 'Processing' tab at the top of the screen.
Since March 2018, the NMR Jeol spectra are supported (only the Free Induction Delay) within the JDF format created from the Jeol Delta software.
- See the Spectral pre-processing for 1D NMR section.
- See a complete example with a JEOL spectra set (JDF Format) - (PDF online)
Since March 2019, the NMR RS2D spectra are supported (both Free Induction Delay and Spectrum in frequency domain) within the SPINit format created from the SPINit software.
- See the Spectral pre-processing for 1D NMR section.
- See a complete example with a RS2D spectra set (SPINit Format) - (PDF online)
Since January 2018, the nmrML format are supported (only the Free Induction Delay).
- See the Spectral pre-processing for 1D NMR section.
- See an example of nmrML convertion of a JEOL spectra set (JDF Format) before uploading within NMRProcFlow - (PDF online)
References
Schober D, Jacob D, Wilson M, Cruz JA, Marcu A, Grant JR, Moing A, Deborde C, de Figueiredo LF, Haug K, Rocca-Serra P, Easton J, Ebbels TMD, Hao J, Ludwig C, Günther UL, Rosato A, Klein MS, Lewis IA, Luchinat C, Jones AR, Grauslys A, Larralde M, Yokochi M, Kobayashi N, Porzel A, Griffin JL, Viant MR, Wishart DS, Steinbeck C, Salek RM, Neumann S. (2018) Anal Chem doi: 10.1021/acs.analchem.7b02795